xray-thc
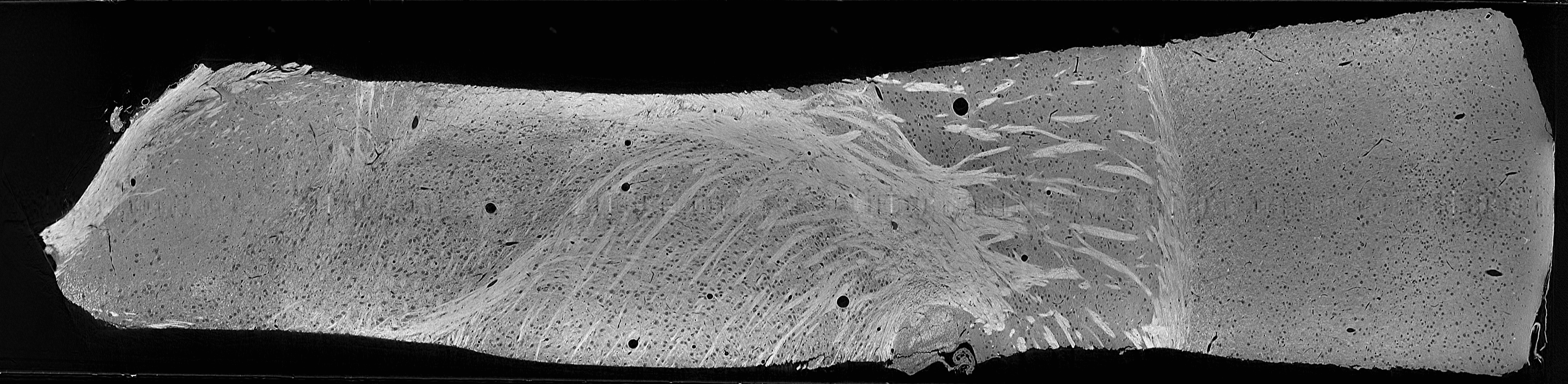
Thalamocortical sample imaged with X-ray microCT
A three-dimensional X-ray microtomography thalamocortical dataset for characterizing brain heterogeneity : Preprint, Data
Python notebooks for pulling down the data from BossDB, along with an instructional README, are provided in the “data_access_notebooks” directory. See the Requirements below needed to use BossDB for data access.
Getting Started
If you’d like to get started, check out our Python notebook for analyzing the microstructure in different ROIs, and generating the analyses provided in Figure 2 and 3 in the paper.
Dataset Description
This dataset consists of a 3D brain volume, generated via microCT, spanning from hypothalamus to cortex. The dataset has dimension 720x1420x5805 (z,y,x), with a 1.17um isotropic voxel volume. The brain areas available are Cortex, Striatum, TRN, VP, Zona Incerta, Internal Capsule, Hypothalamus, and Corpus Callosum. Human-annotated ground truth data are available for both brain area classification and samples for microstructure segmentation of 4 brain areas.
Relevant notebooks:
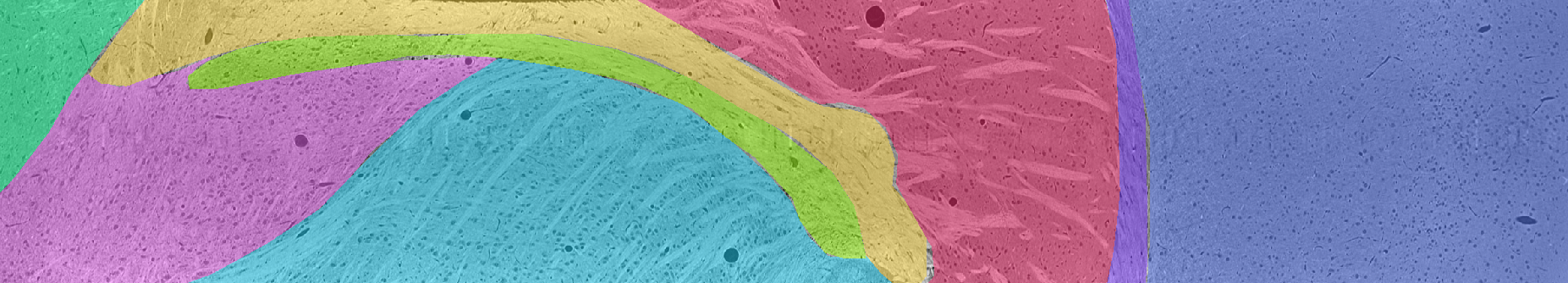
Brain Area Annotations
Brain area annotations are available for eight different regions of interest. Complete human annotations of brain area are available for slices z = 109, 159, 209, 259, 309, 359, 409, 410, and 460. Thus, across each of these 9 slices, every pixel is labeled with the following values: Clear Label = 0, Cortex = 1, Striatum = 2, TRN = 3, VP = 4, Zona Incerta = 5, Internal Capsule = 6, Hypothalamus = 7, Corpus Callosum = 8.
Relevant notebooks:
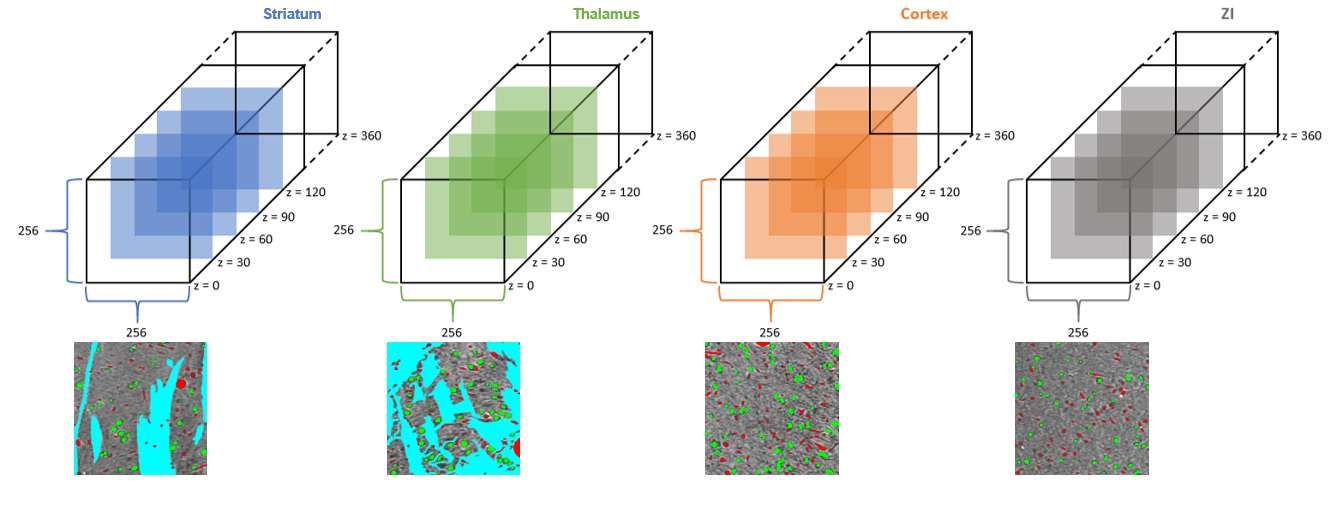
Pixel-level Microstructure Annotations
Microstructure segmentation (of cell bodies, blood vessels, and myelinated axons) are available for 4 regions of interest: Cortex, Striatum, Thalamus (mostly VP and some TRN), and Zona Incerta. For each of these 4 regions, there is a 256x256x360 (x,y,z) volume available for which slice z (0 indexed) = 30, 60, 90, 120, 150, 180, 210, 240, 270, 300, and 330 have been densely annotated.
Microstructures were annotated with the following values: background = 0, cells = 1, blood vessels = 2, myelinated axons = 3.
Relevant notebooks:
Requirements
General requirements to run code associated with this project are Python 3.x and Jupyter Notebook. Additionally, to pull down the data from bossDB, we make use of blosc, intern, and numpy, all of which can be installed via pip.
Alternatively, to download all dependancies, navigate to your local version of this repo via the command line and run
pip install -r requirements.txt
For further details about our versions of specific packages, please check requirements.txt
Team
- Aishwarya H. Balwani (AishwaryaHB)
- Eva L. Dyer (evadyer)
- William Gray Roncal (wrgr)
- Erik C. Johnson (erikjohnson24)
- Joseph D. Miano (jmiano)
- Judy A. Prasad